PCR: The Basic Theory
The capacity of the technique called the polymerase chain reaction (PCR) to amplify many million-fold any known DNA fragment from a complex mixture in a short time has revolutionized all areas of the life sciences, making it one of the most widely used molecular techniques in use today.
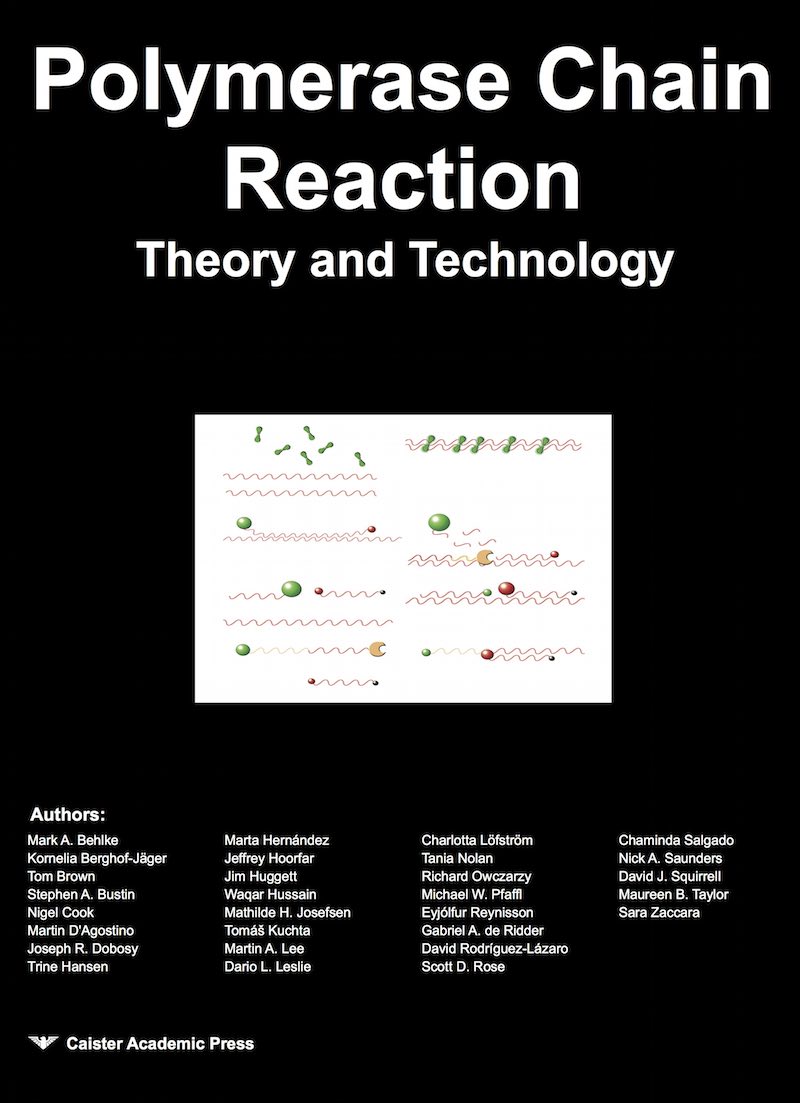
Quite simply, PCR is a laboratory technique used to makes a huge number of copies of a piece of DNA. In other words it is a way of amplifying DNA or increasing the amount of a specific piece of DNA. The core principle of PCR is the use of an enzyme called DNA polymerase to make a copy of a DNA strand. Normally DNA exists as a double strand, but the enzyme can only work on a single strand. Therefore it is first necessary to separate the strands of DNA. This is done by applying heat.
Applying heat to DNA denatures the double strand to single strands. In the first PCR step the double strand melts to single stranded DNA, a step known as denaturation. The enzyme can work on the single strand to make a copy. However, the enzyme needs a small region of double strand to get started. So it is necessary to add a short piece of single strand DNA, called the primer , that binds specifically to a particular place on the single strand molecule. This binding, or annealing, is achieved by cooling the PCR mixture again. This step in the PCR process is called annealing.
Then the DNA polymerase enzyme gets to work and copies the single strand molecule, starting at the bound primer region. This final step, extension, is so-called because the DNA extends the DNA from the annealed primer and makes a complementary copy of the single strand. In practice, two different primers are used. A forward primer that binds to one strand and a reverse primer that binds to the opposite strand. The choice of primers is important as each primer binds the DNA in a specific place. The only DNA that is copied is the region between the forward primer binding location and the reverse primer binding location.
Both single strands of the original double-stranded DNA molecule are copied. Therefore the result is two double stranded molecules each identical to the original double-stranded DNA fragment.
The three PCR steps are repeated for around 30 or 40 cycles. Each cycle doubles the number of double stranded DNA molecules.
Normally PCR is performed on a machine called a PCR thermal cycler or PCR machine. The PCR thermal cycler rapidly heats and cools the PCR reaction mixture thus allowing the denaturation, annealing and extension to occur.
PCR Problems
In practise, the PCR technique is more complicated than outlined above and it is not always simple to achieve the desired result. For example, primers can bind non-specifically to multiple places on the DNA strand. This results in a mixture of amplified product. In addition, various factors such as impurities in the DNA mixture, can prevent the reaction occurring at all. For further information see our section on PCR TroubleshootingReal-Time PCR
Conventional gel-based PCR, or legacy PCR, as described above, has several disadvantages: it is labour-intensive, not easily automated or adapted for high throughput applications, and quantification of nucleic acids is arduous. The introduction of novel reagents, chemistries and instrumentation platforms has modified legacy PCR to create a new technology, real-time, fluorescence-based quantitative PCR (qPCR also known as real-time PCR or qRT-PCR) that addresses these issues. The use of real-time PCR allows monitoring of the progress of a PCR reaction in real time using fluorescent reporter molecules included in the PCR mixture. For further information see our section on Real-Time PCRFurther reading
- Real-Time PCR: Advanced Technologies and Applications
- Real-Time PCR in Food Science: Current Technology and Applications
- Quantitative Real-time PCR in Applied Microbiology